TA Cloning Overview
TA cloning is a form of subcloning that uses complementary base pairing and ligation to join a single insert and a backbone. First, the backbone, called a T-vector, must be configured as a linear vector with single base pair (bp) 3' thymine (T) overhangs. Many T-vectors have already been constructed and are available for purchase from biotechnology companies. On the other hand, you can construct T-vectors manually using restriction enzymes.
One way uses restriction enzymes that leave single bp 3' T overhangs as sticky ends after cutting. Alternatively, you can cut the vector using restriction enzymes that leave blunt ends and add 3' T overhangs. Mixing the cleaved vector with dideoxythymidine triphosphate (ddTTP) and a terminal transferase enzyme adds single bp 3' T overhangs to the blunt ends.
Next, allowing for hybridization, the insert must be constructed as a linear DNA sequence with single bp 3' adenine (A) overhangs. This process can be done very simply through PCR using Taq polymerase. Due to the terminal transferase activity of Taq polymerase, single bp 3' A overhangs are added to the ends of the amplified product. After the backbone and insert are constructed, they mix and hybridize due to the 3' T and A overhangs. Then, the backbone and insert join using a DNA ligase to produce the final cloning vector.
The image below shows a general overview of TA cloning. An insert with single bp adenine (A) overhangs on the 3'-ends, and a vector with single bp thymine (T) overhangs on the 3-ends. After mixing, through hybridization and ligation, they join to make a final cloning vector. 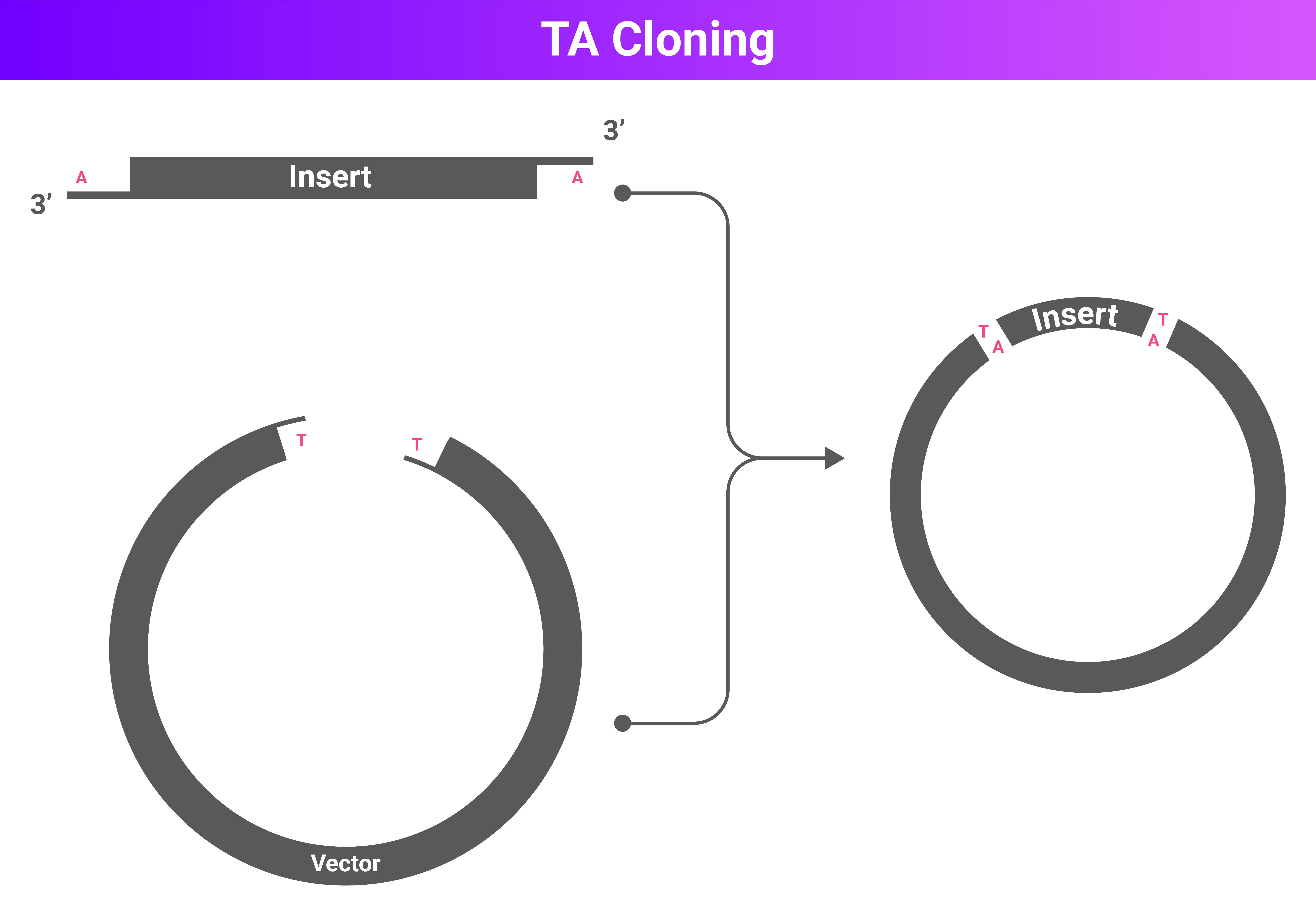
TA Cloning Tutorial
With Mendelgen, performing a simulation of TA cloning is straightforward. Follow the steps outlined below.
- Open Mendelgen, https://mendelgen.com/, and sign in.
- Select and click a vector from your library to be used as the backbone.
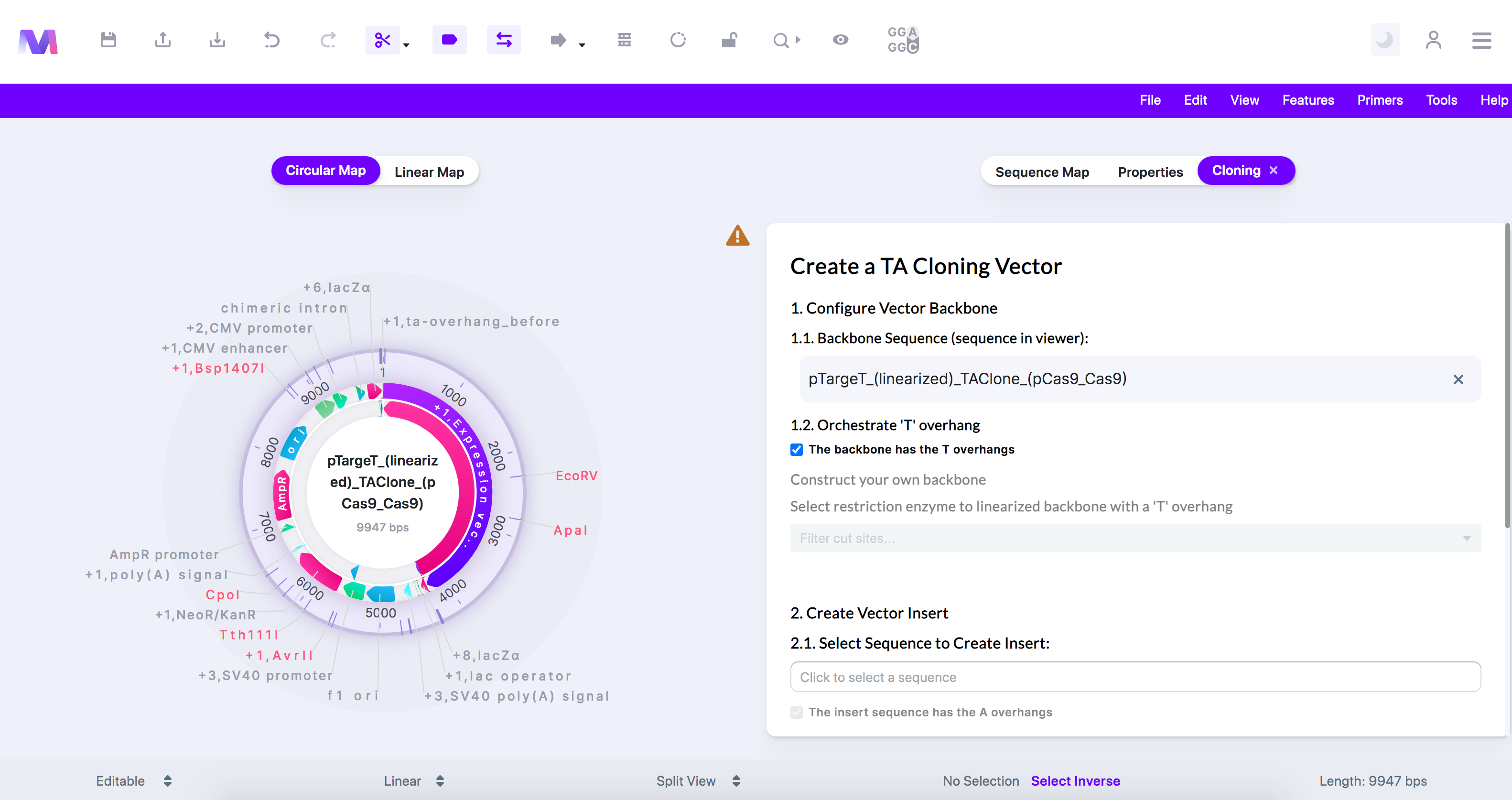
- When you open the vector page, your selected backbone will show in the viewer.
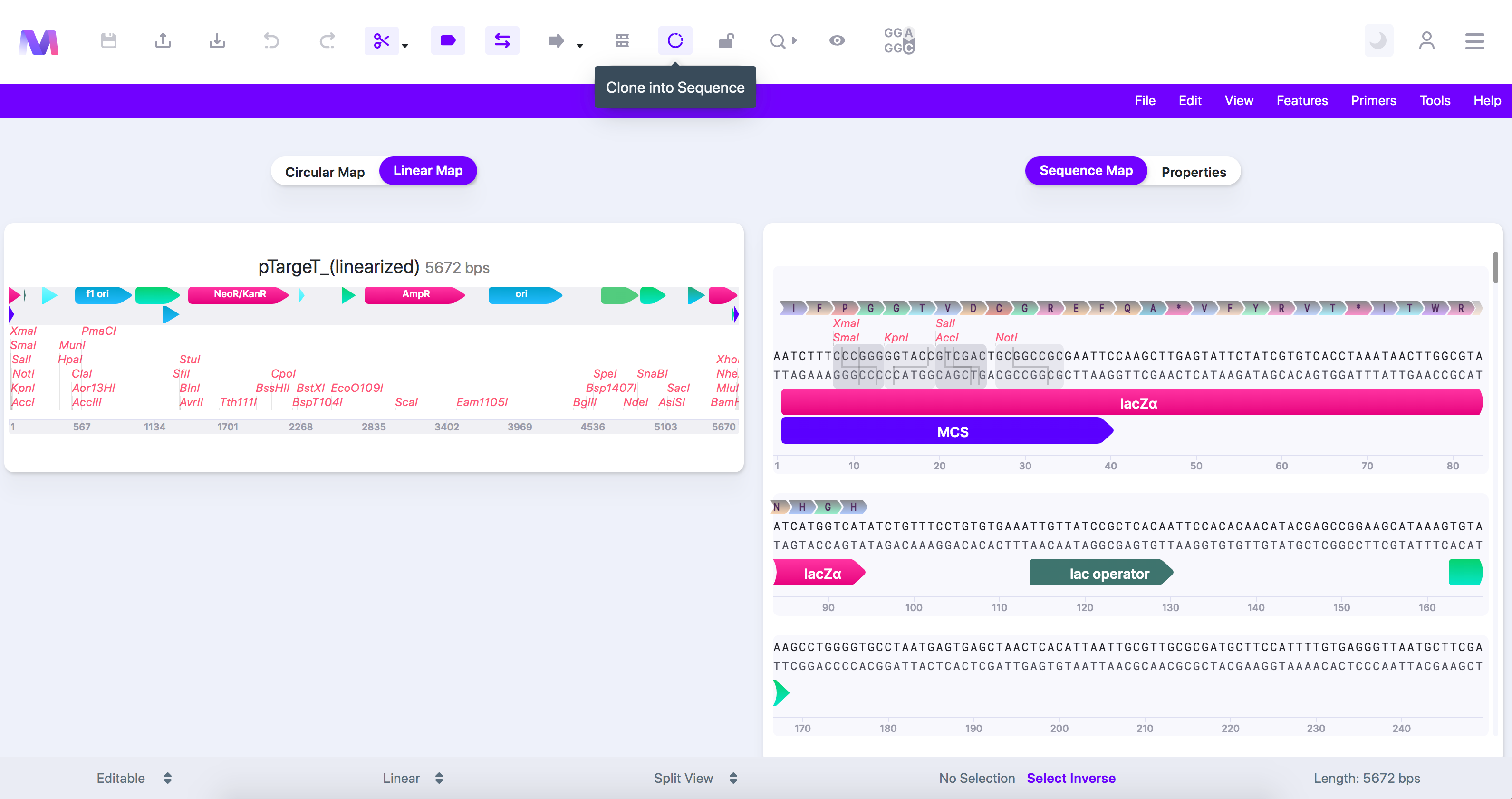
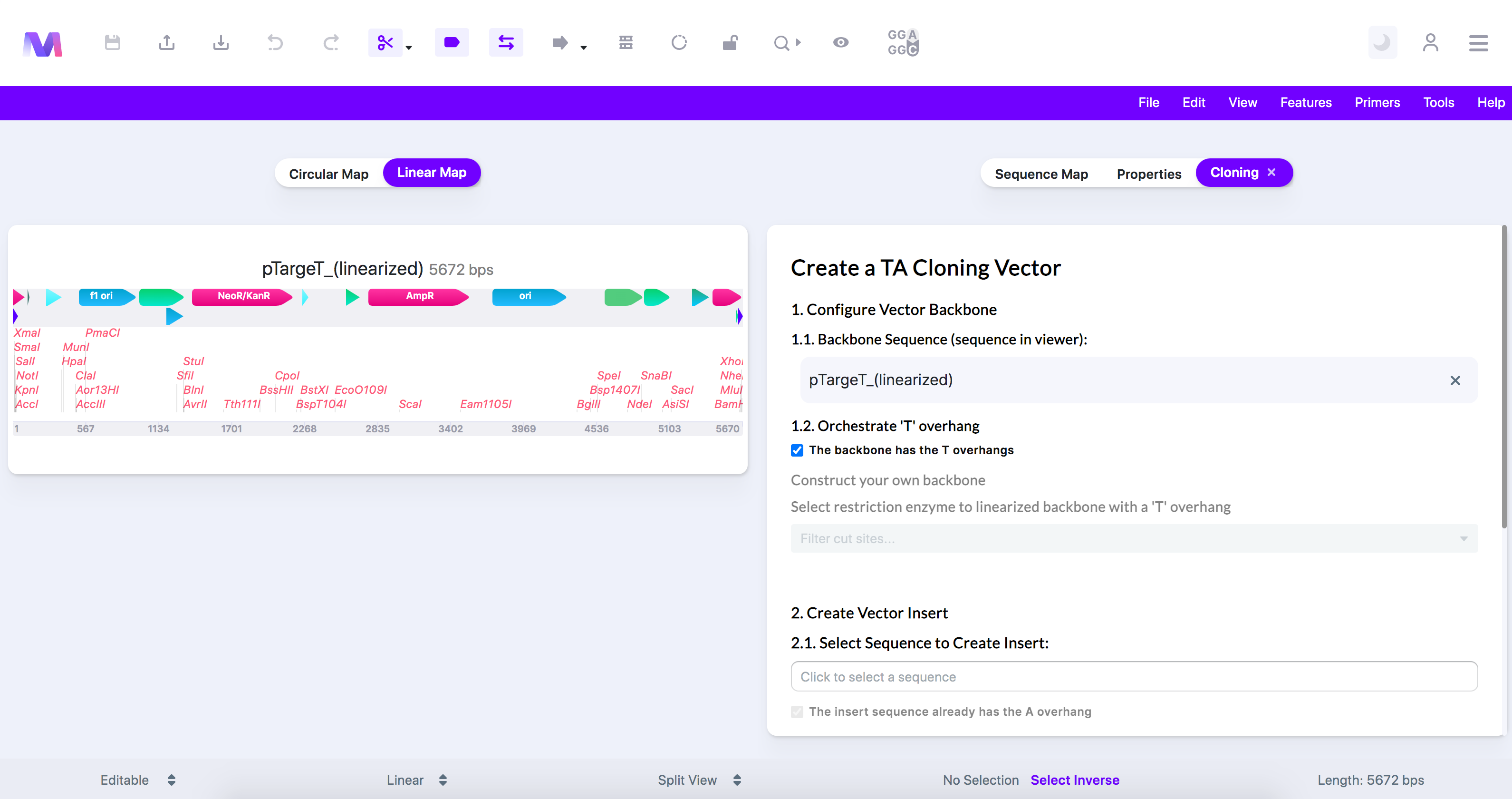
- We have chosen the T-vector 'pTargetT (linearized)' in this example as it is from a TA cloning kit and already contains the 'T' overhangs.
- Click the cloning tool button, displayed as a circular DNA sequence, in the toolbar at the top of the page.
- Select 'TA Cloning' in the dropdown.
- A tab titled 'Create a TA Cloning Vector' will open to the viewer's right.
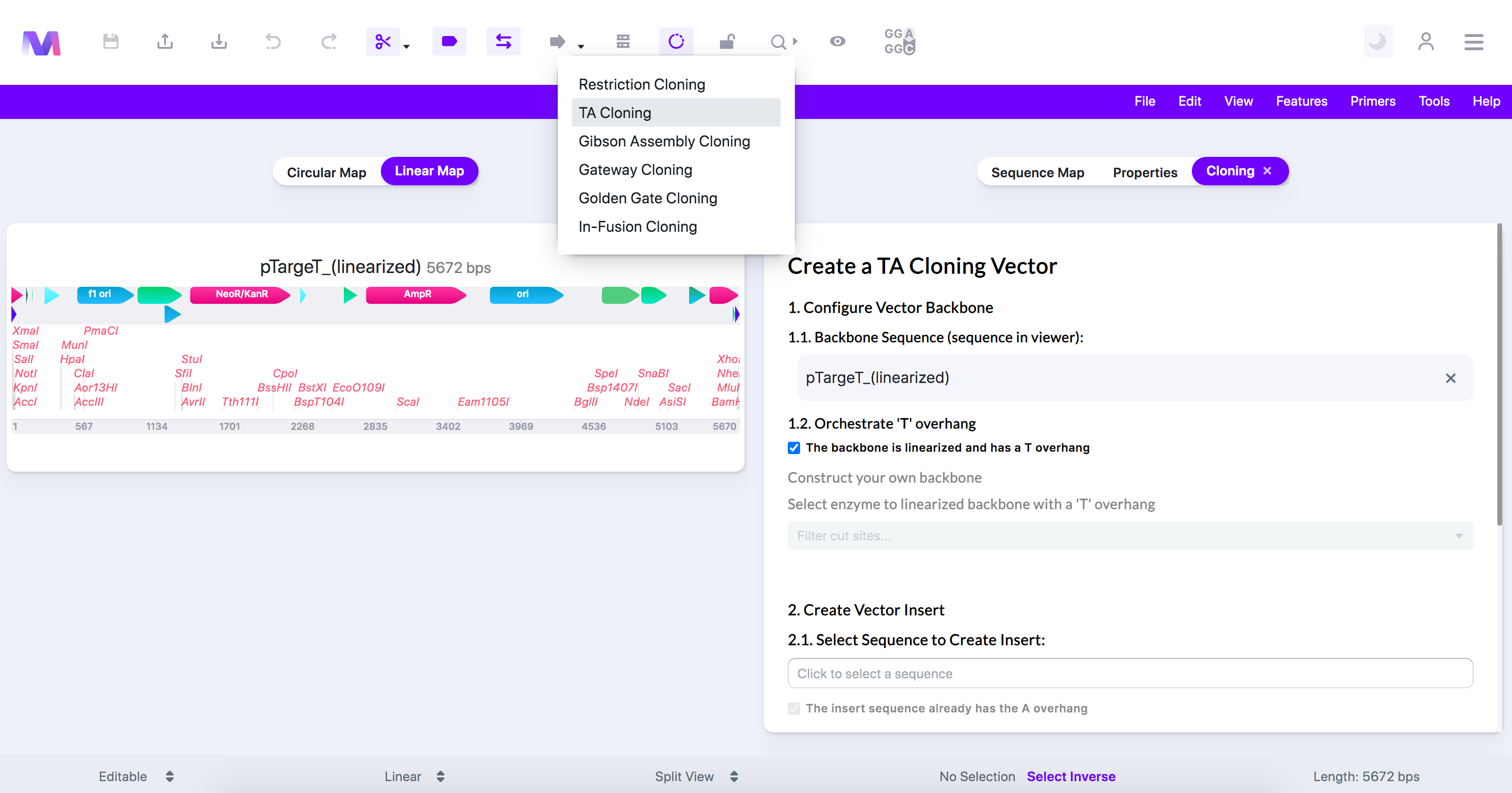
- A tab titled 'Create a TA Cloning Vector' will open to the viewer's right.
- Configure the backbone to have the 'T' overhangs necessary in TA cloning.
- The backbone is the sequence displayed in the viewer.
- Select if the backbone has been linearized and already has the 3' 'T' overhangs by clicking 'The backbone has the 3' T overhangs.'
- If the backbone has the 'T' overhangs, as shown below, move to the next step.
- If the backbone does not have the 'T' overhangs, do the following.
- Select a restriction enzyme to linearize the backbone.

- If the enzyme produces blunt ends, then:
- Choose one or two enzymes that produce one or two cuts.
- The 'T' overhangs get added in the simulation.
- If the enzyme produces sticky ends:
- You can choose one or two enzymes, and two cuts must occur.
- The sticky ends must be single bp 3' 'T' overhangs.
- Select a restriction enzyme to linearize the backbone.
- If the backbone has the 'T' overhangs, as shown below, move to the next step.
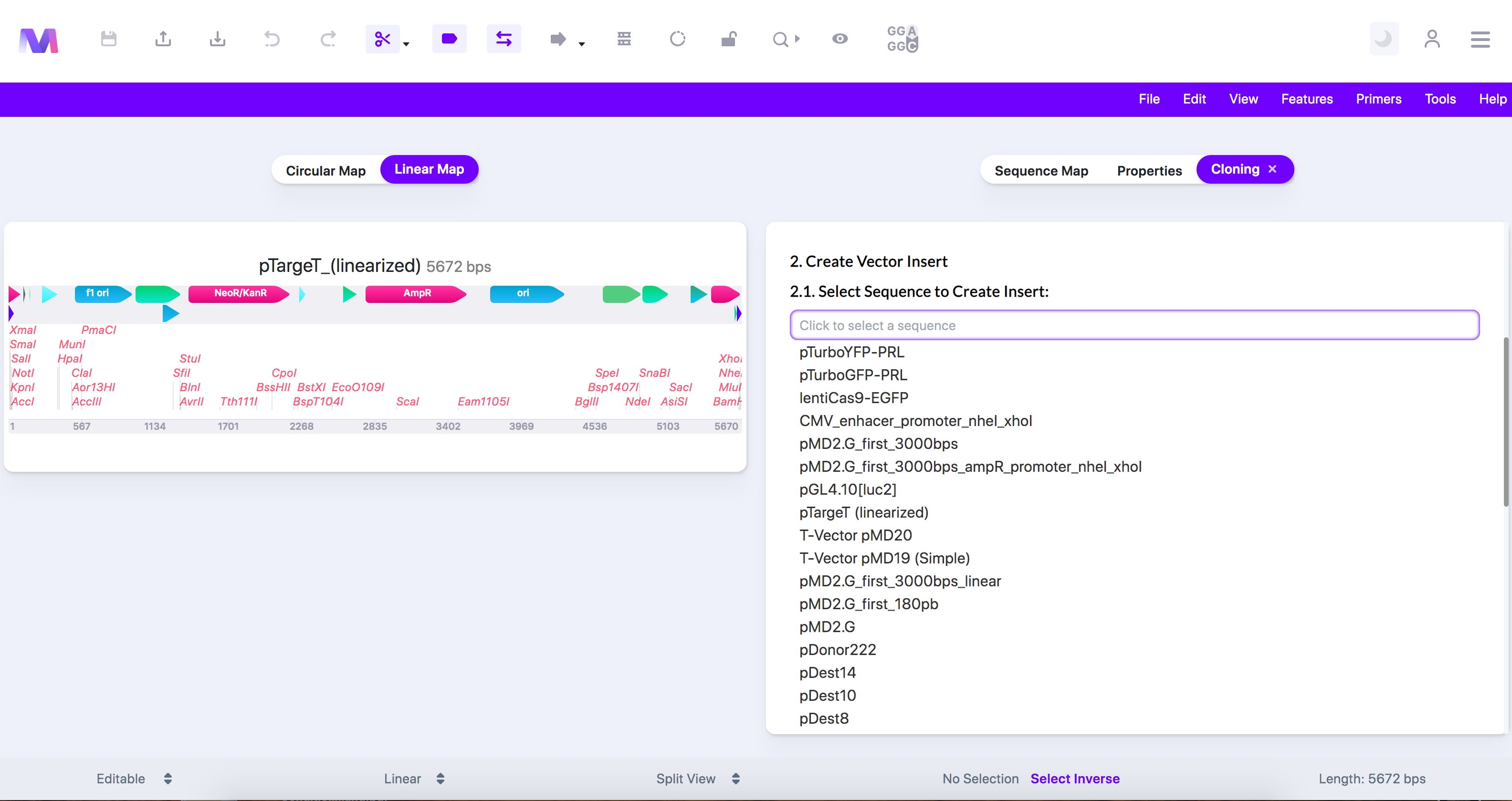
- Select and configure the insert sequence containing the gene(s) of interest.
- Select a sequence loaded from your library.
- If your sequence is linearized and already has the 3' 'A' overhangs check the box by clicking 'The insert sequence has the 3' A overhangs.'
- If the insert has the 'A' overhangs, move to the next step.
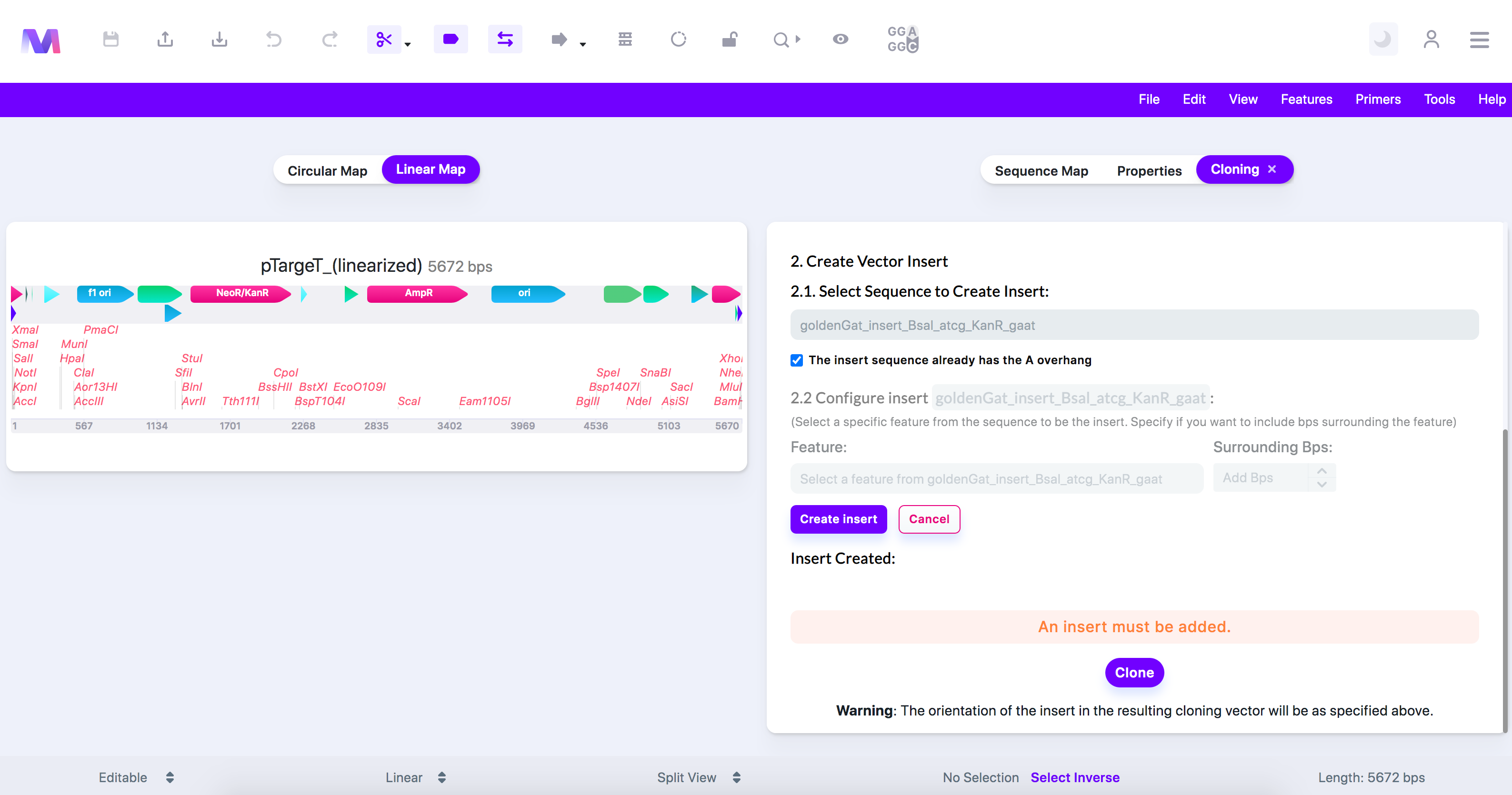
- If the insert does not have the 'A' overhangs, do the following where you can make further configurations.
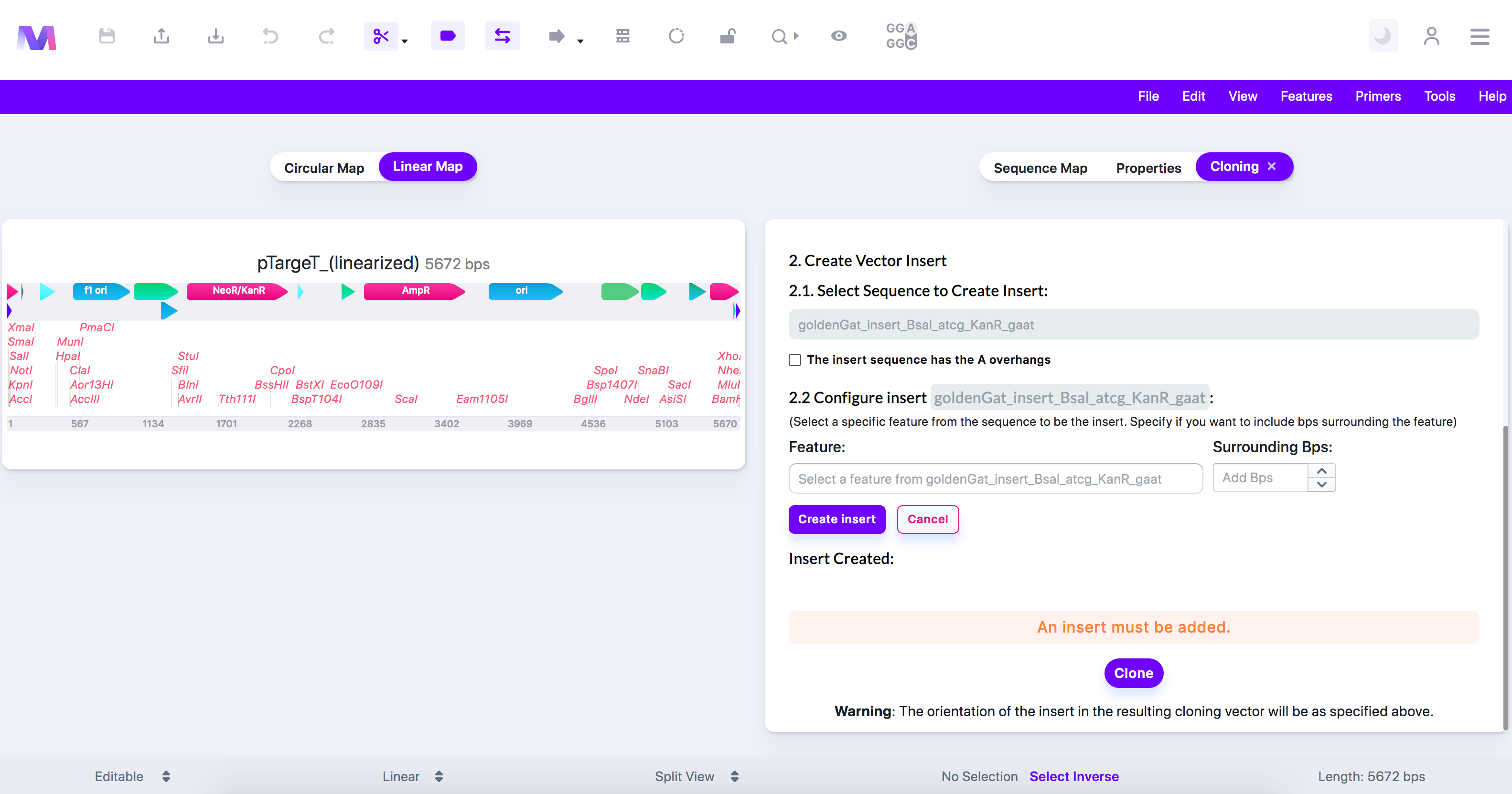
- Under 'Configure Insert,' specify only the region of interest you want to use as the insert. You can skip this step if you wish to insert the entire sequence.
- Choose a feature on the insert and specify if you want to include any base pairs surrounding the feature.
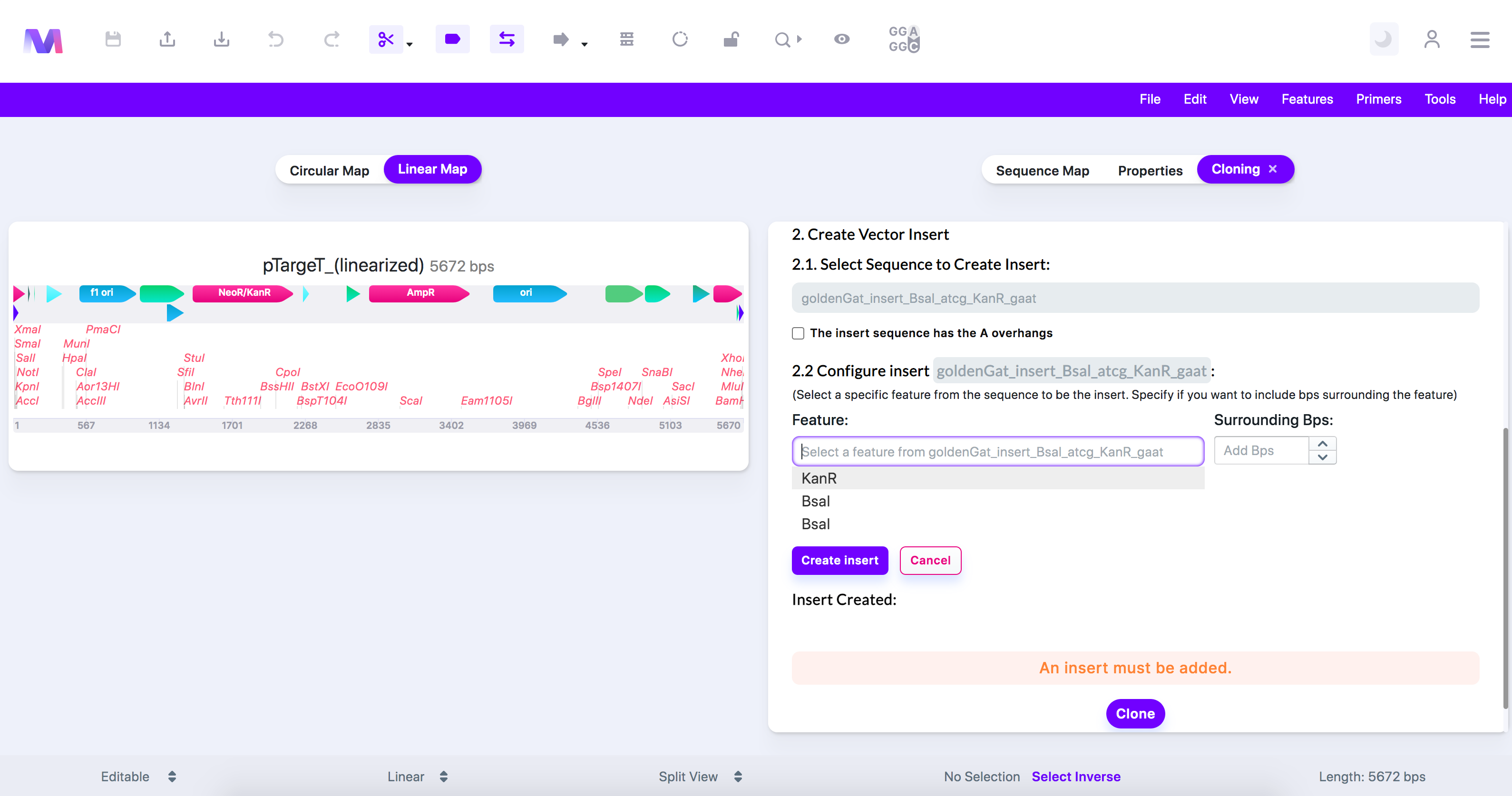
- PCR will occur in the simulation, and the 'A' overhangs will get added to the insert.
- If the insert has the 'A' overhangs, move to the next step.
- Click 'Create Insert,' and the insert will get added under 'Insert Created.'
- Note that you can only add one insert.
- Select a sequence loaded from your library.
- Check that the correct insert was added under 'Insert Created.'
- Click the 'X' by the sequence under 'Insert Created' to remove the insert from the list so you can add a different insert.
- Insert shown under 'Insert Created.'
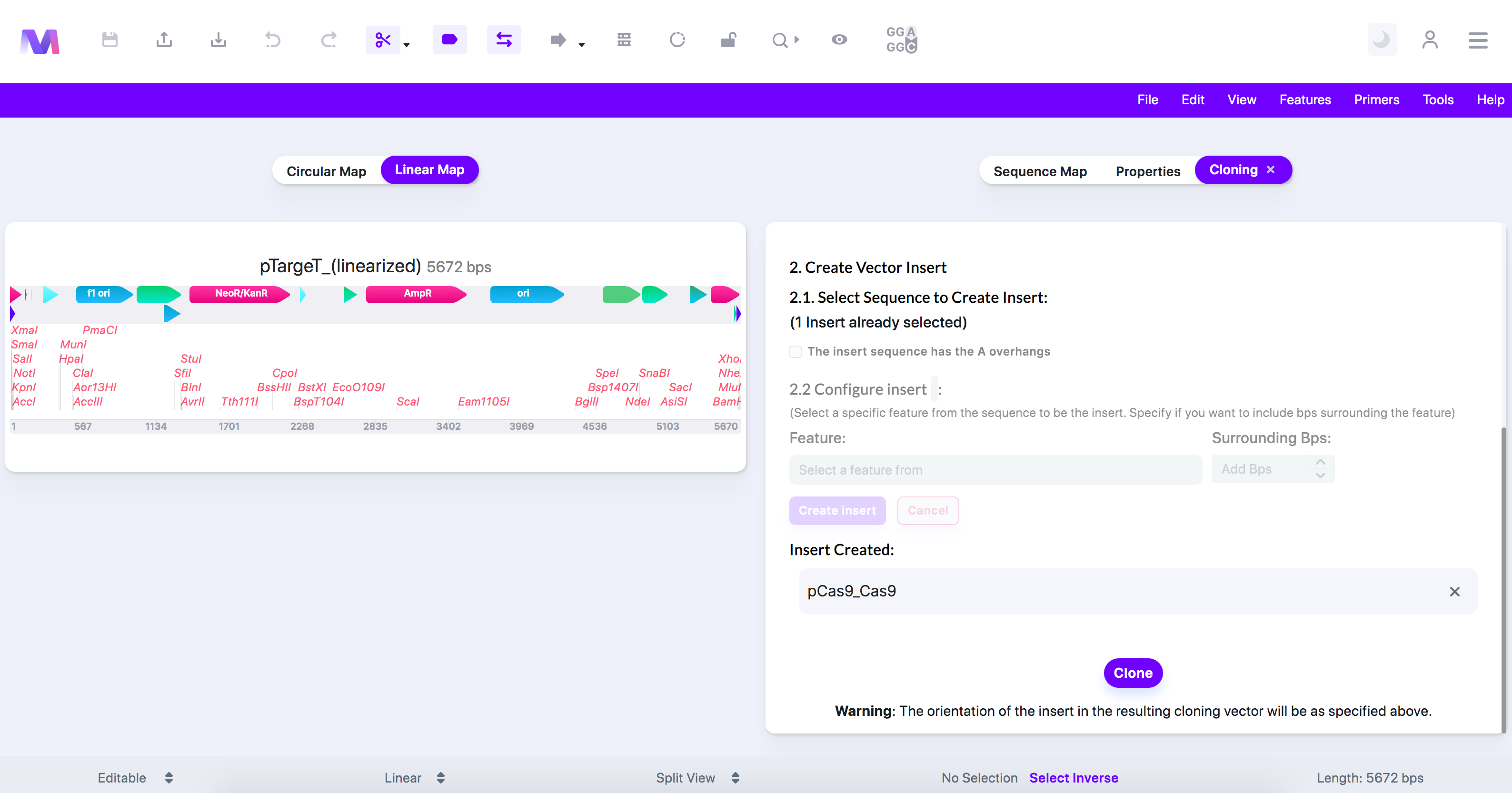
- Click the 'Clone' button to perform a TA Cloning simulation.
- You will not be able to click the 'Clone' button if any errors exist.
- The image below shows an error highlighted above the cloning button preventing cloning.
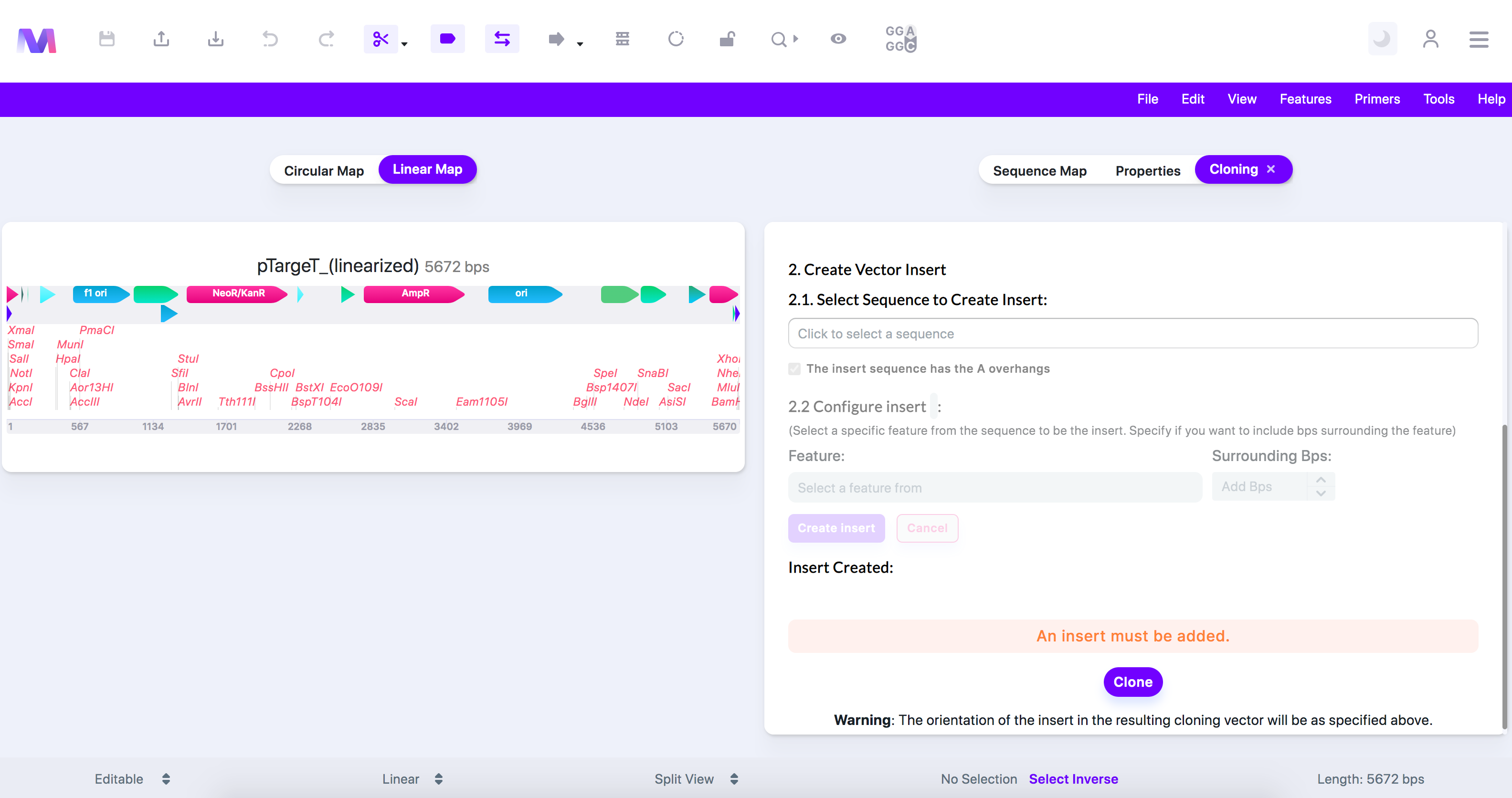
- Errors and their descriptions:
- An Insert must be added.
- Too many inserts have been added. Only 1 insert is allowed.'
- The backbone and insert cannot have the same name.
- The sequence name used as the backbone cannot be identical to the insert. You can construct the insert using the same sequence as the backbone, but you must select a feature.
- The backbone '(backbone name provided)' must be linearized if it already contains the 3' 't' overhangs.
- If you supply a backbone that already has 'T' overhangs, then it must be linear so that TA cloning can occur.

- If you supply a backbone that already has 'T' overhangs, then it must be linear so that TA cloning can occur.
- If the backbone already has the 'T' overhangs, the first bp must be 'A,' but it is '(bp provided).'
- The 'T' overhangs on the backbone are on the 3' end. Therefore, the first bp on the 5' end must be an 'A.'
- If the backbone already has the 'T' overhangs, the last bp must be a 'T,' but it is '(bp provided).'
- The 'T' overhangs on the backbone are on the 3' end. The last bp, which is on the 3' end, must be a 'T.'
- A restriction enzyme must be selected to cut the backbone.
- If you are constructing your T-vector from the backbone, you must provide a restriction enzyme to cut the backbone, creating places for the 'T' overhangs to be added.
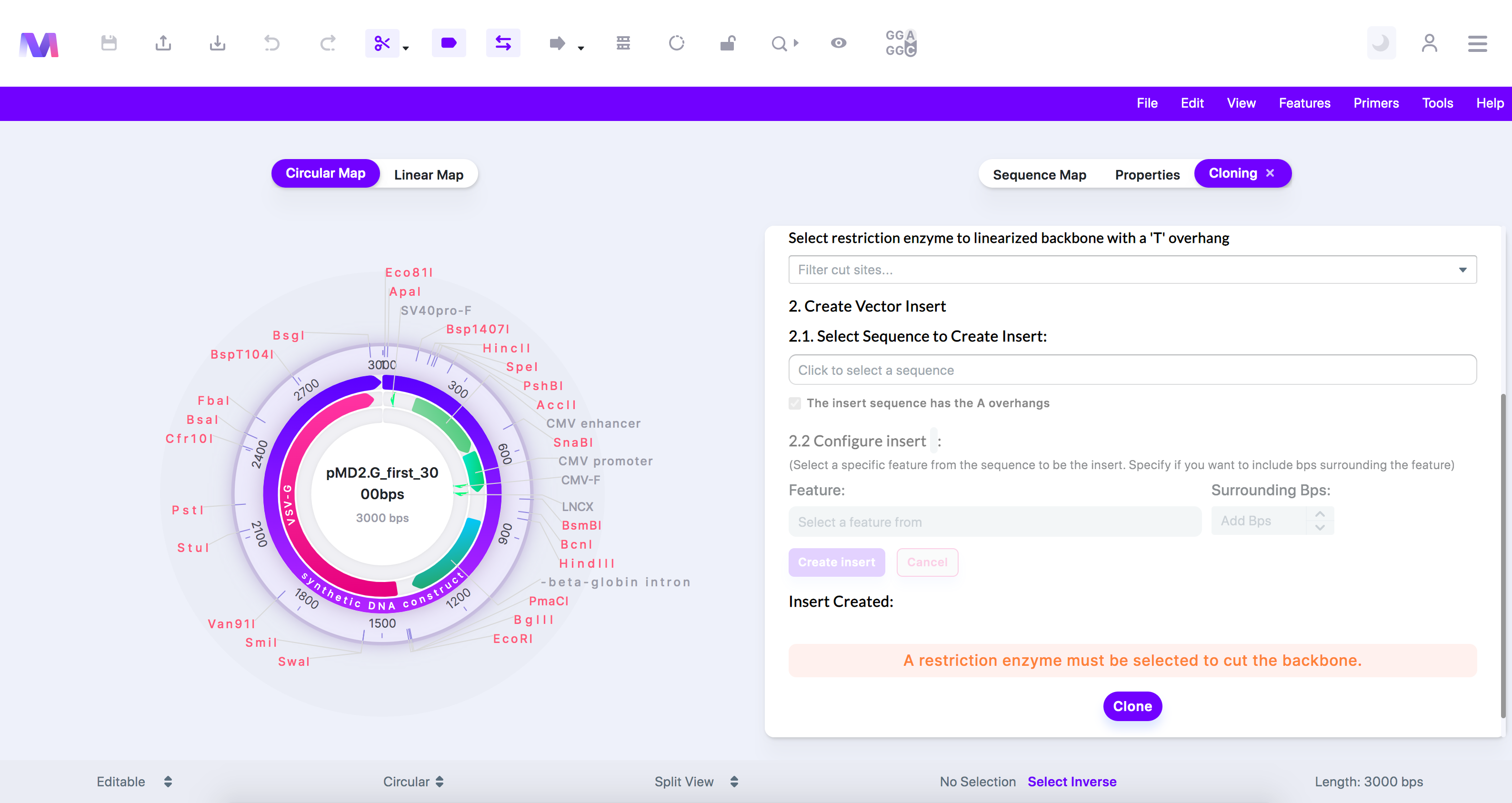
- If you are constructing your T-vector from the backbone, you must provide a restriction enzyme to cut the backbone, creating places for the 'T' overhangs to be added.
- Too many restriction enzymes have been selected. Only 2 are allowed.
- When constructing your T-vector from the backbone you cannot have more than two restriction enzymes because too many fragments will be created, and we will not be able to discern which fragment will get used as the T-vector.
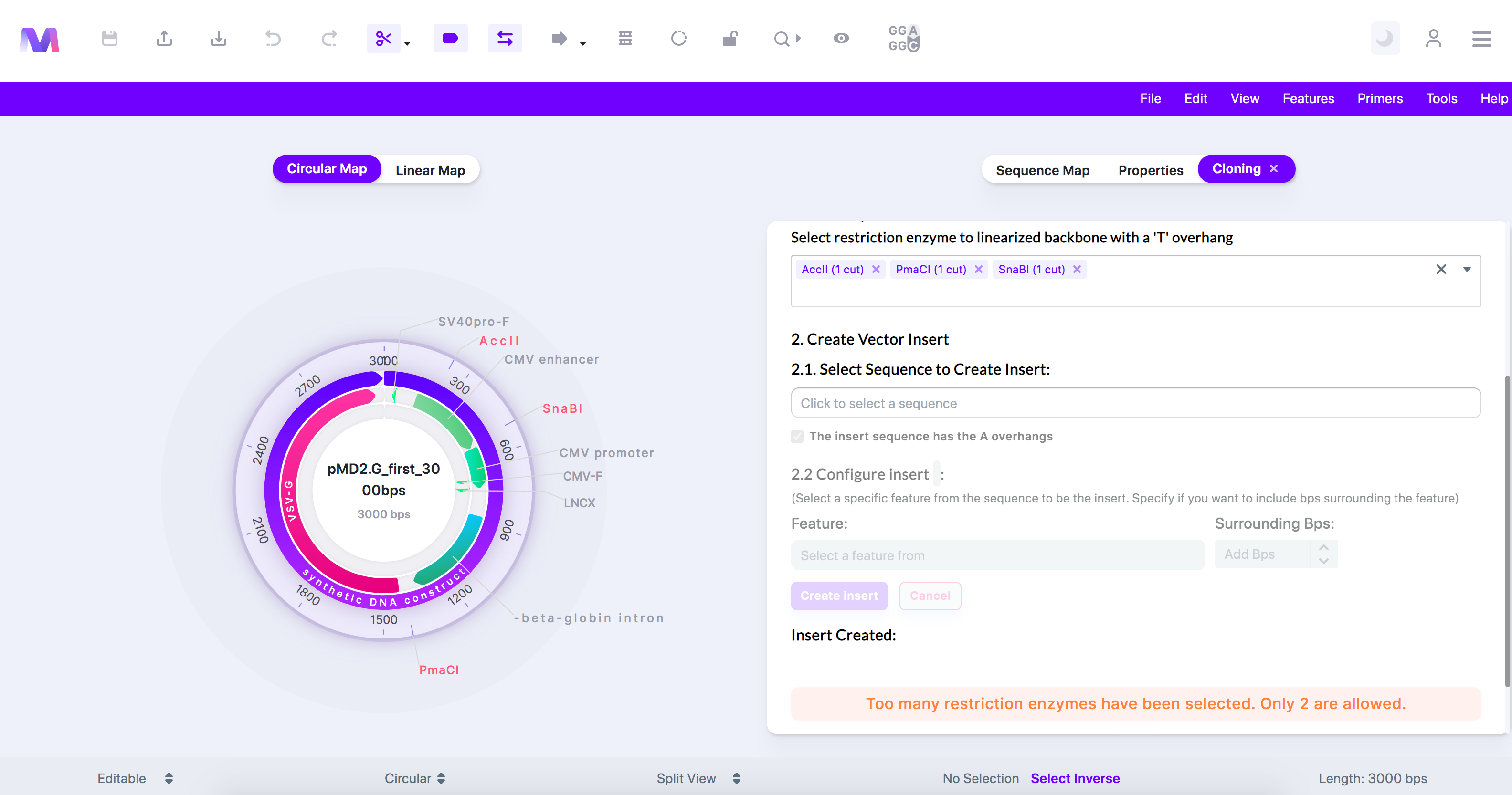
- When constructing your T-vector from the backbone you cannot have more than two restriction enzymes because too many fragments will be created, and we will not be able to discern which fragment will get used as the T-vector.
- The backbone must have at least 1 cut.
- If you are constructing your T-vector from the backbone, you must provide at least one cut to the backbone, creating places for the 'T' overhangs to be added.
- The backbone cannot be cut more than twice.
- When constructing your T-vector from the backbone, you cannot have more than two cuts because too many fragments will be created, and we will not be able to discern which fragment will get as the T-vector.
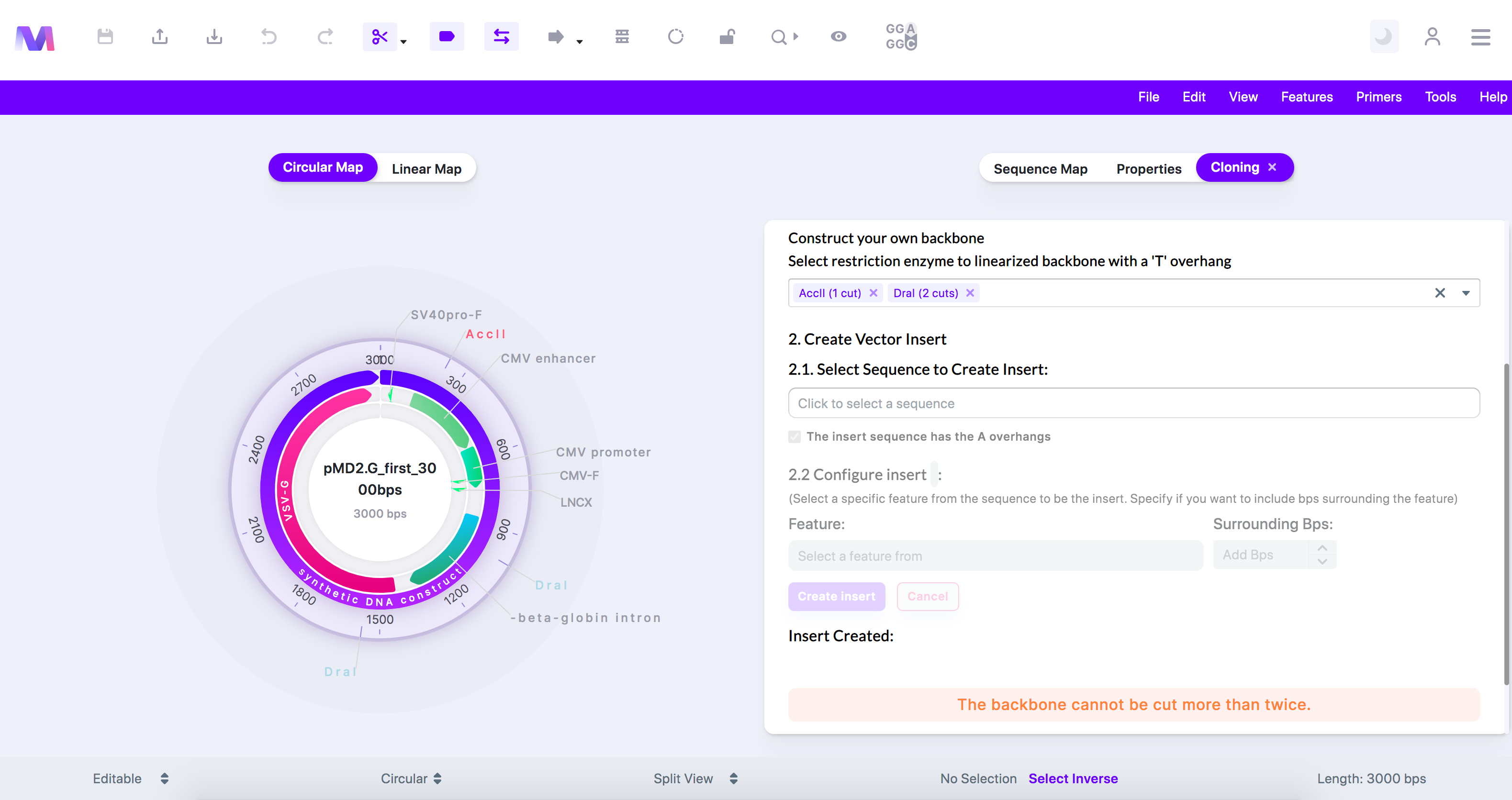
- When constructing your T-vector from the backbone, you cannot have more than two cuts because too many fragments will be created, and we will not be able to discern which fragment will get as the T-vector.
- The enzyme '(enzyme name provided)' producing blunt ends cannot be selected, while the enzyme '(enzyme name provided)' producing overhangs is selected.
- When constructing a T-vector from the backbone, you cannot select enzyme-producing blunt ends, and enzyme-producing overhangs simultaneously. This is because the differences in the ends would prevent the 'T' overhangs from being produced correctly.
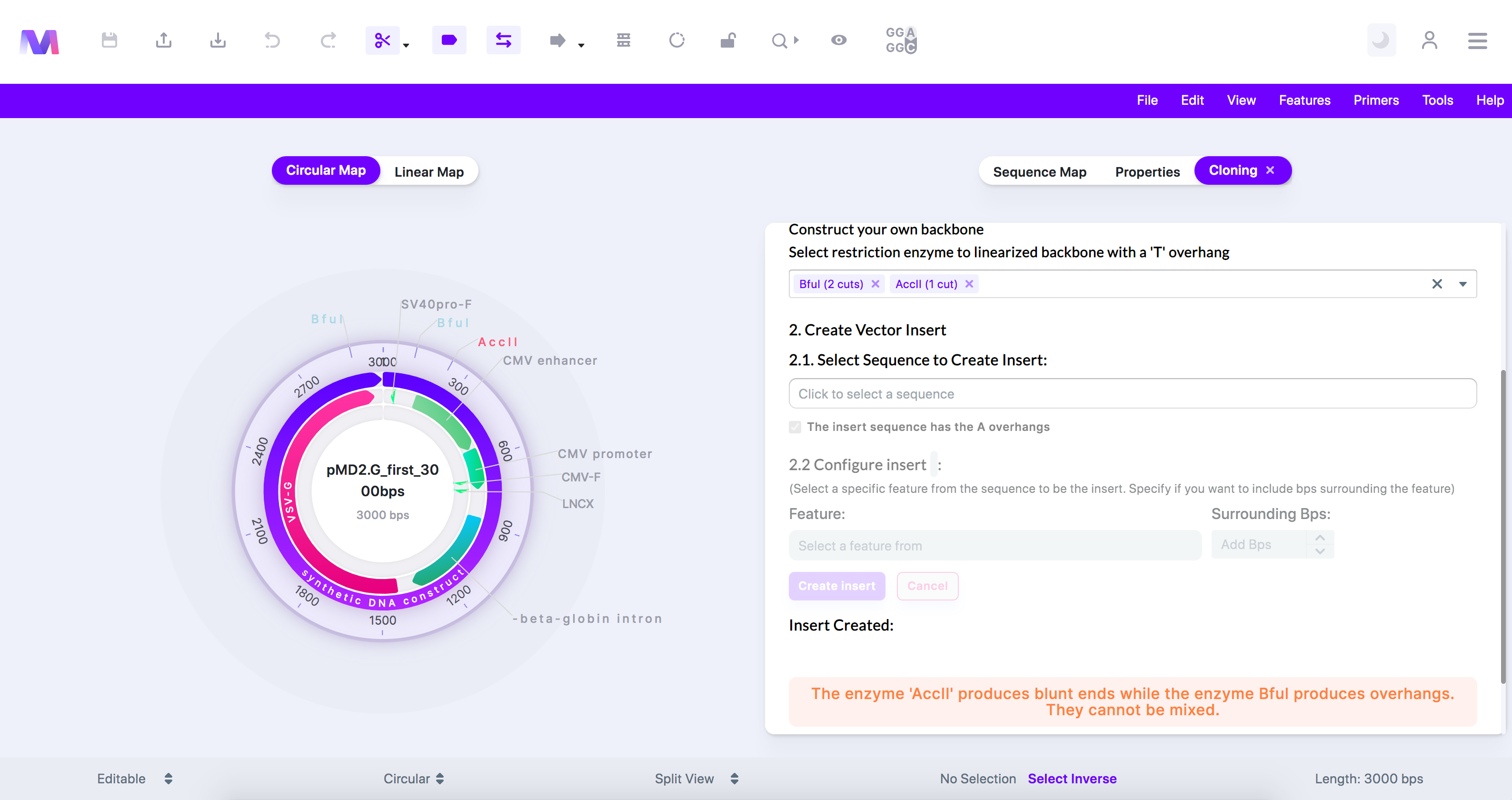
- When constructing a T-vector from the backbone, you cannot select enzyme-producing blunt ends, and enzyme-producing overhangs simultaneously. This is because the differences in the ends would prevent the 'T' overhangs from being produced correctly.
- The enzyme '(enzyme name provided)' produces 1 cut with overhangs. There must be 2 cuts when the cuts produce overhangs.
- There must be two cuts when using restriction enzymes that produce sticky ends. The 'T' overhangs come from the cuts; therefore, you need two cuts to create a T-vector with 3' 'T' overhangs on both sides.
- The enzyme '(enzyme name provided)' produces a 5' overhang, which will not work in TA cloning.
- In TA cloning, all the overhangs occur on the 3' ends.
- The 3' 'T' overhangs will not be constructed correctly. The enzyme '(enzyme name)' produces an overhang with the bps '(bps provided)'.
- The T-vector created from the backbone will not have 3' 'T' ends because the restriction enzyme selected produces an overhang with other bps.
- The 3' 'T' overhangs will not be constructed correctly. The enzyme '(enzyme name provided)' does not produce a 3' 'T' overhang.
- The T-vector created from the backbone will have one correct 3' 'T' overhang, but the other 3' 'T' overhang will not be constructed correctly due to the restriction enzyme chosen.
- The insert '(insert name provided)' must be linear if it already contains the 'A' overhangs.
- An insert that already has 'A' overhangs must be linear so it can be used directly in TA cloning.
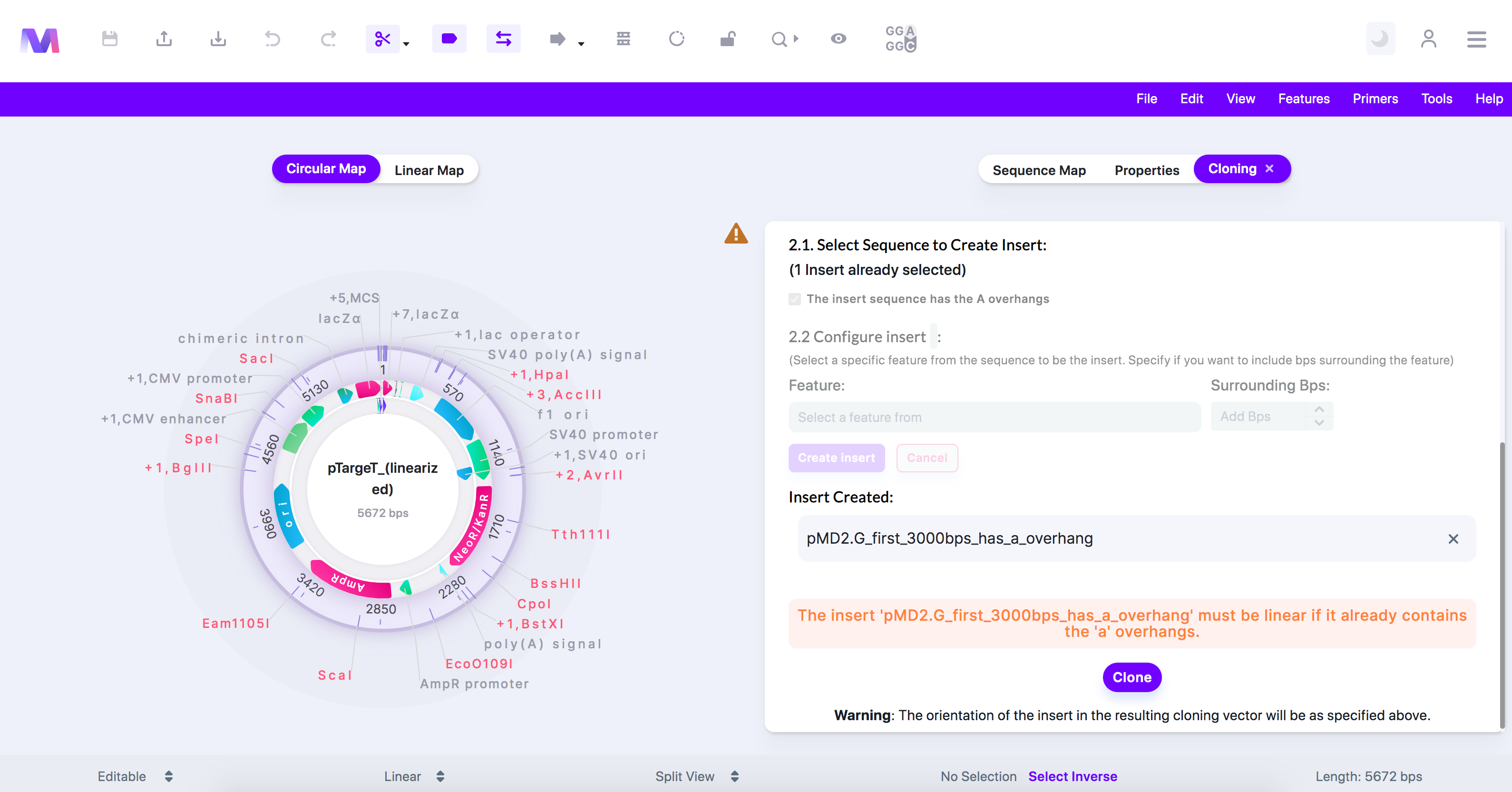
- An insert that already has 'A' overhangs must be linear so it can be used directly in TA cloning.
- If the insert '(insert name provided)' already has 3' 'A' overhangs, the first bp must be 'T,' but it is '(bp provided).
- The 'A' overhangs on the backbone are on the 3' end. The first bp on the 5' end must be 'T,' so the 3' side will be 'A.'
- If the insert '(insert name provided)' already has the 3' 'A' overhangs, the last bp must be 'A', but it is '(bp provided).
- The 'A' overhangs on the backbone are on the 3' end. The last bp, which is on the 3' end, must be 'A'.
- A feature cannot be selected or bps added if the insert already contains the 'A' overhangs.
- If the 'A' overhangs already exist on the sequence uploaded, you cannot pick a feature from the sequence to create the insert.
- You should now see your final resulting vector in the viewer! 🎉
- Mendelgen has simulated the following.
- Linearization, if needed, of the backbone.
- Addition of 3' 'T' overhangs, if need be, to the backbone.
- If required, PCR amplifies the insert with Taq polymerase, adding the 3' 'A' overhangs.
- Pairing and ligation of the backbone and insert to produce the final vector.
- You can view your finished cloning simulation with the resulting vector in the viewer below.
- Mendelgen has simulated the following.