Advanced Settings
Auto-Annotating Features
Mendelgen enables researchers to auto-annotate the sequences. To do that, select all sequence (or a part of it) and select "Auto-Annotate Features" from the "Tools" tab in the Control Panel.
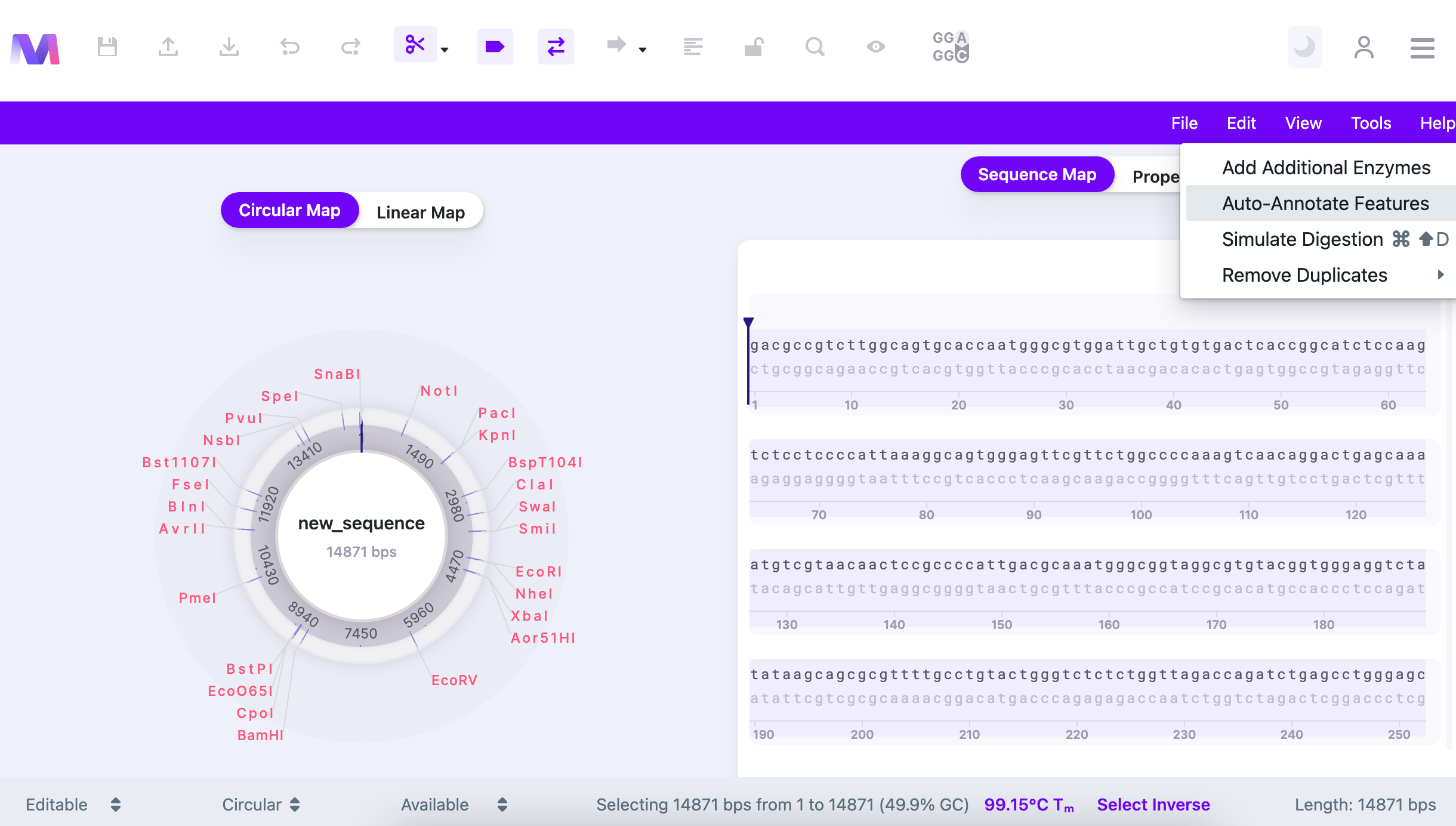
The search will start immediately and the list of detected features will be displayed.
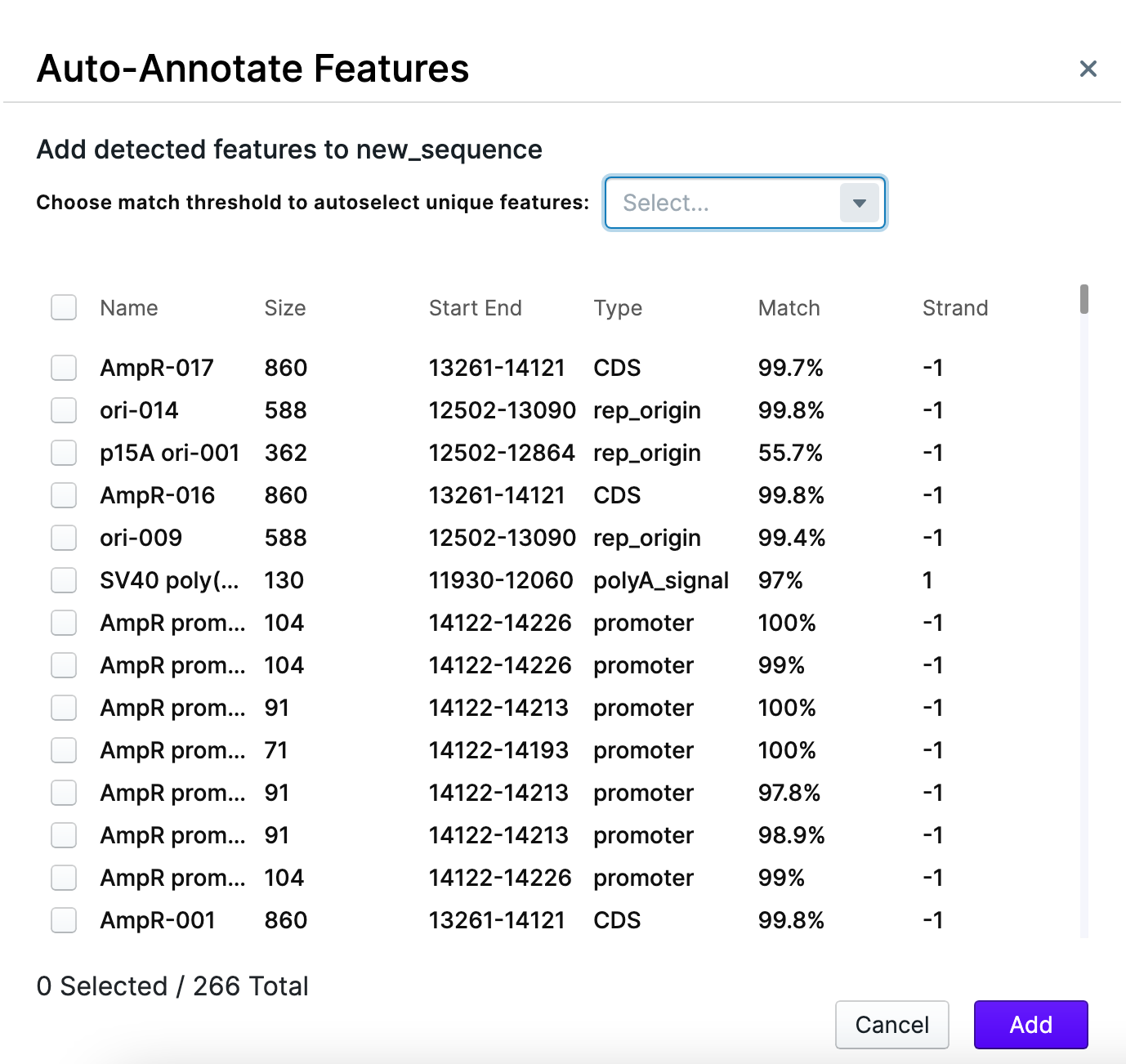
You are able to change the match threshold and pick the desirable features from the list by check-marking them. The match threshold shows the confidence, with which the feature is being recognized. In this example, the highest confidence level (100% match threshold) was selected, and then – required features hand-picked.
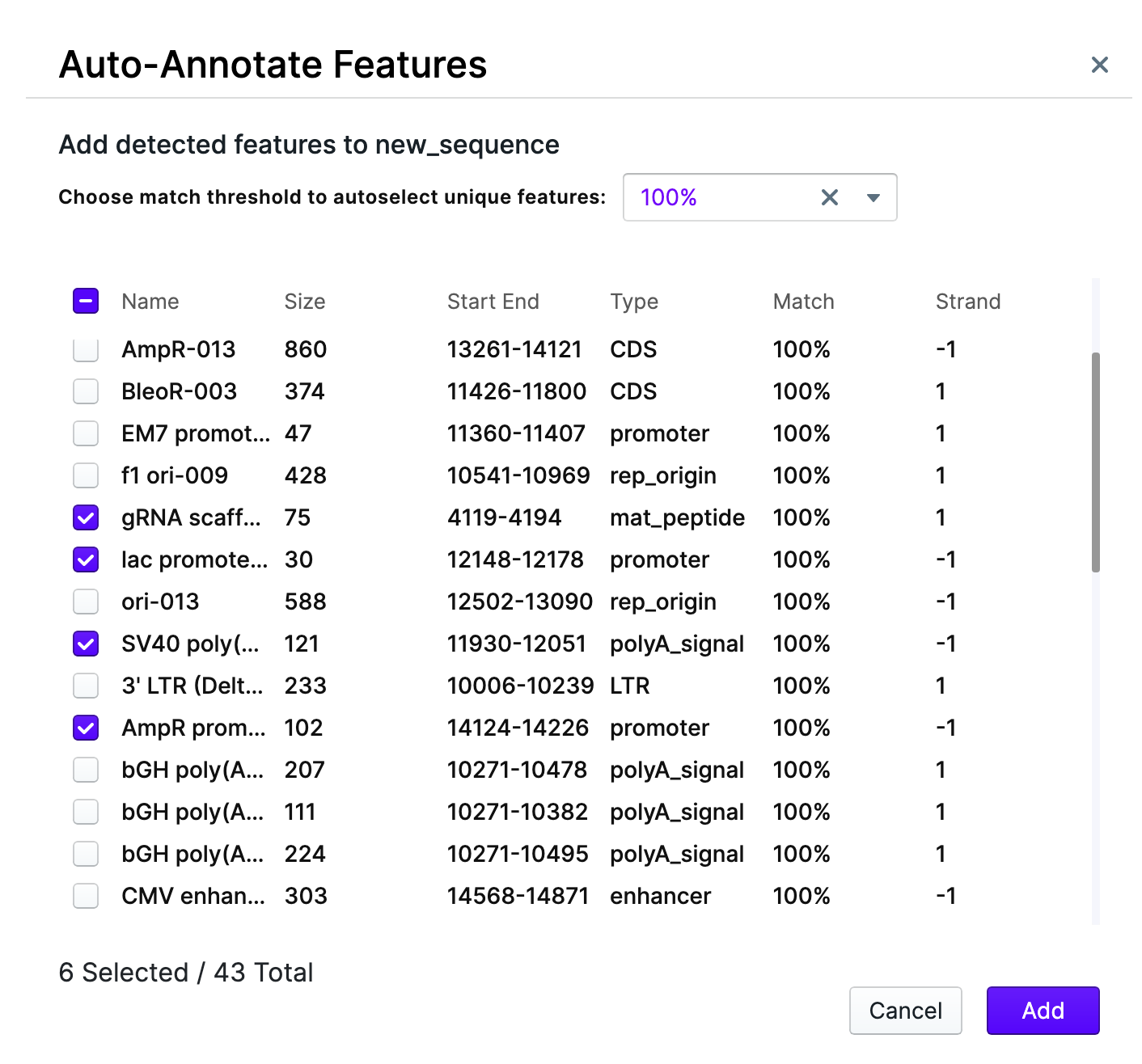
After pressing “Add” the features will be displayed on the sequence map.
Thermodynamic Settings
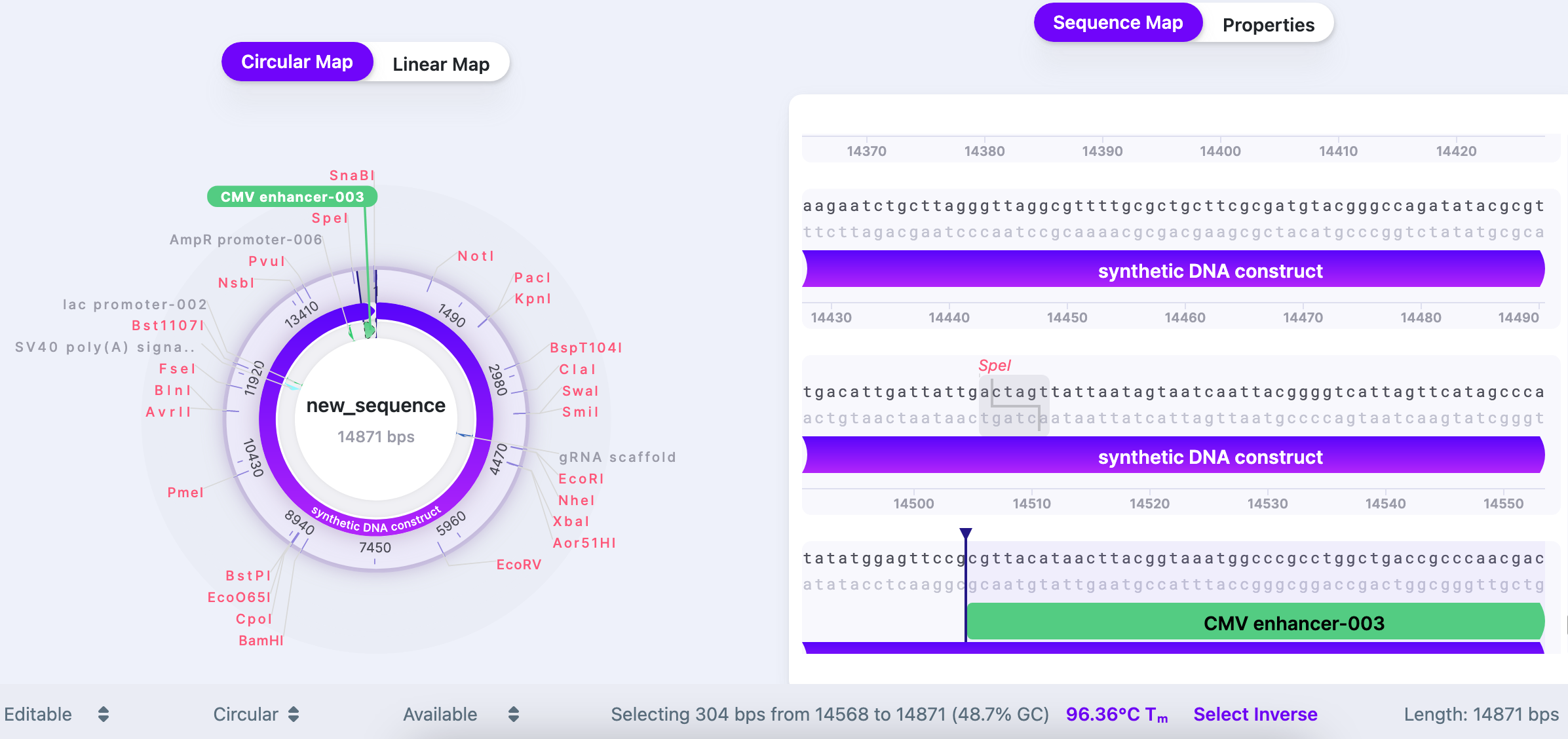
The thermodynamic settings are available from the bottom panel in your workspace upon selection of the sequence.
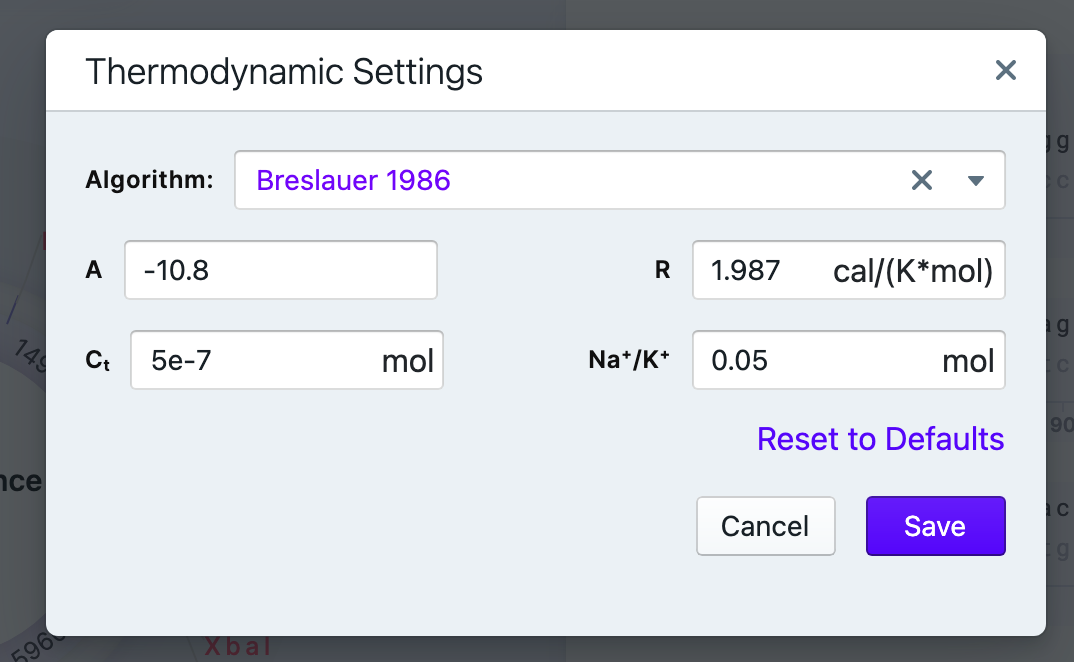
In the Thermodynamic Settings tab you can select the algorithm and modify several parameters (depending on the algorithm you choose): Cp (heat capacity, in mol) and Na+/K+ concentration (in mol), A (constant of -0.0108 kcal K-1 ᐧ mol-1 (accounts for helix initiation during annealing/melting)), R (gas constant of 1.987 cal/(K*mol) (constant that scales energy to temperature)), and Ct (oligonucleotide concentration in mol). For example, as DNA is a polyanionic molecule, salt level affects the melting temperature: at the same temperature, DNA will melt slower under low salt conditions.
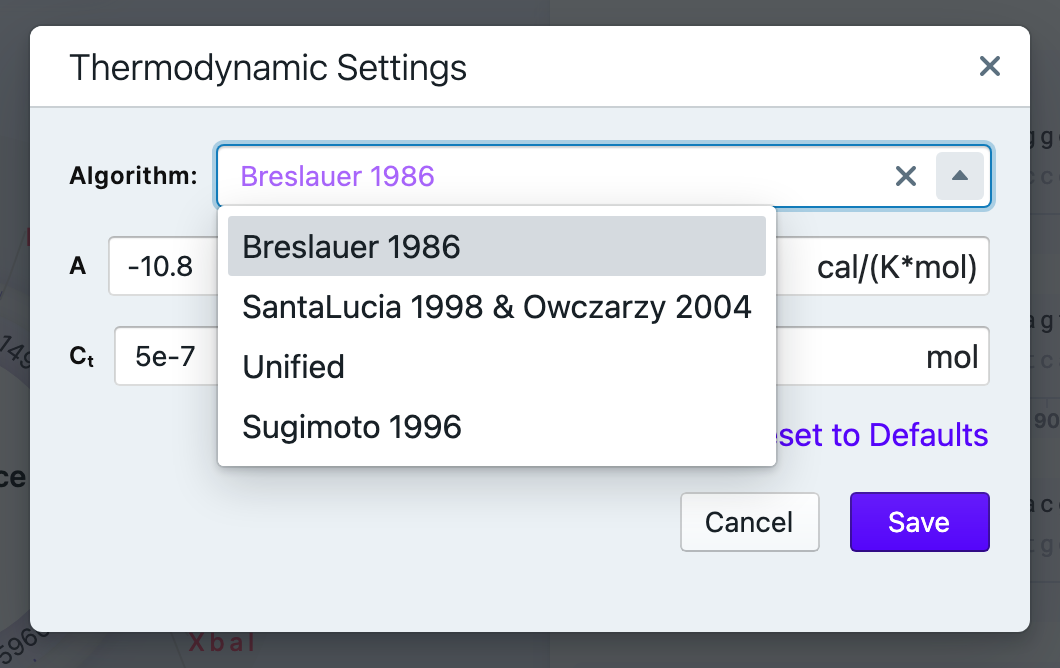
The available algorithms
- Breslauer 1986
- SantaLucia 1998 & Owczazry 2004
- Unified
- Sugimoto 1996
You can always reverse the changes by resetting the thermodynamic settings to defaults.
Adding Additional Enzymes
Mendelgen’s default list contains the most common enzymes, however, you may add the less common enzymes from an extended list.
To access the feature, you have two options:
1) Select the “Show/Hide Cut Sites” button on the Control Panel. In the dropdown menu, the top option is “Add Additional Enzymes”.
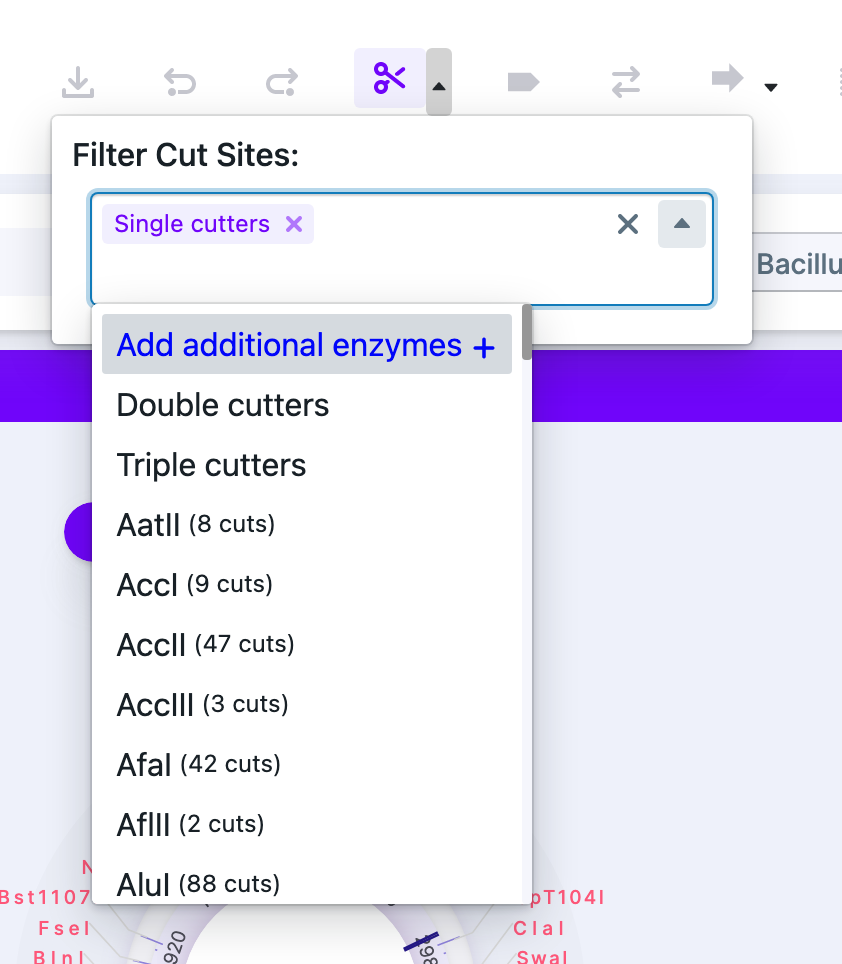
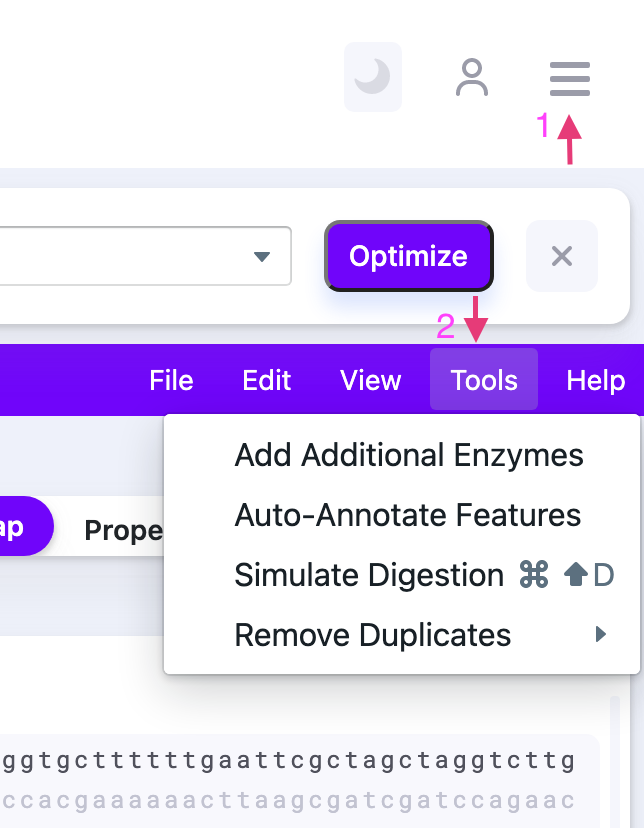
2) Select “Add Additional Enzymes” from the “Tools” tab in the control panel.
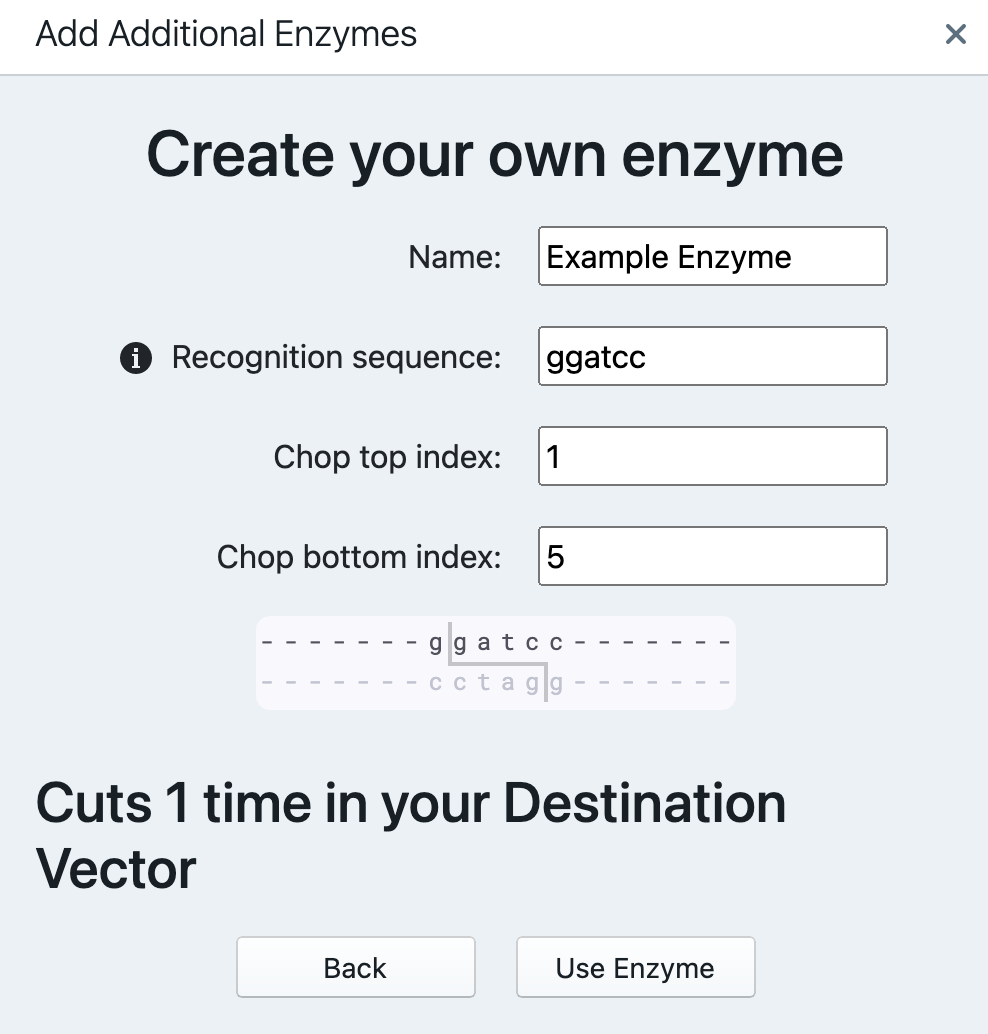
By clicking “Add Additional Enzymes” the window pops up.
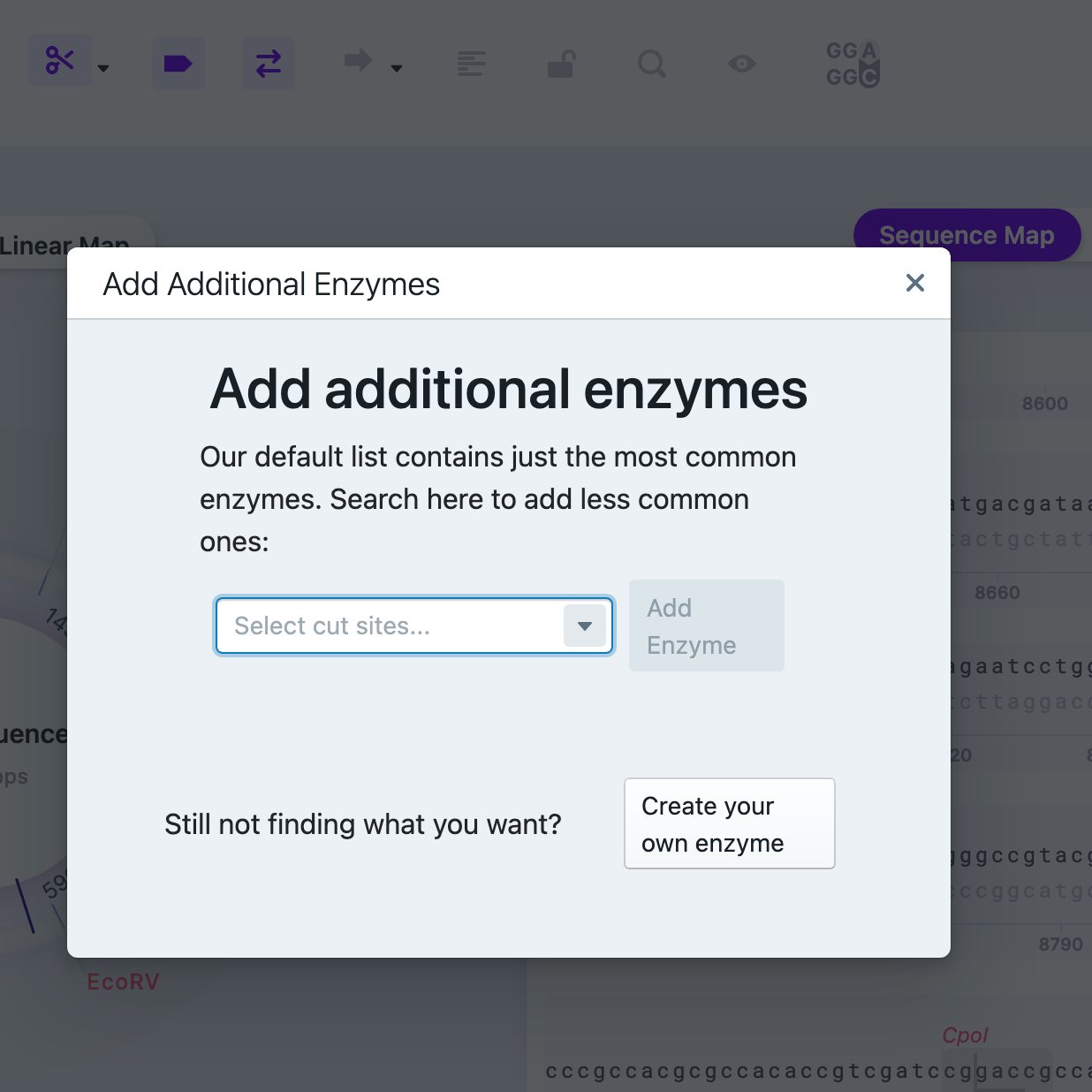
The dropdown menu contains a list of less common restriction enzymes. You may add more manually by pressing “Create Your Own Enzyme”. To fill it out, you need to provide name, recognition sequence (minimum of 4 bps long), chop index (top and bottom) – after what nucleotide the enzyme will cut the sequence (counting from the left). You will be able to see how many times this enzyme will cut your vector (displays values between 0 and 10 (“More than 10”). After finalizing, press “Use Enzyme”.